SNBL Software & Docs
Pylatus: beamline control software
What is Pylatus
Pylatus is beamline control software for diffractometers based on Dectris Pilatus detectors and ICEPAP motor controls originally developed for the BM01 beamline at SNBL@ESRF.
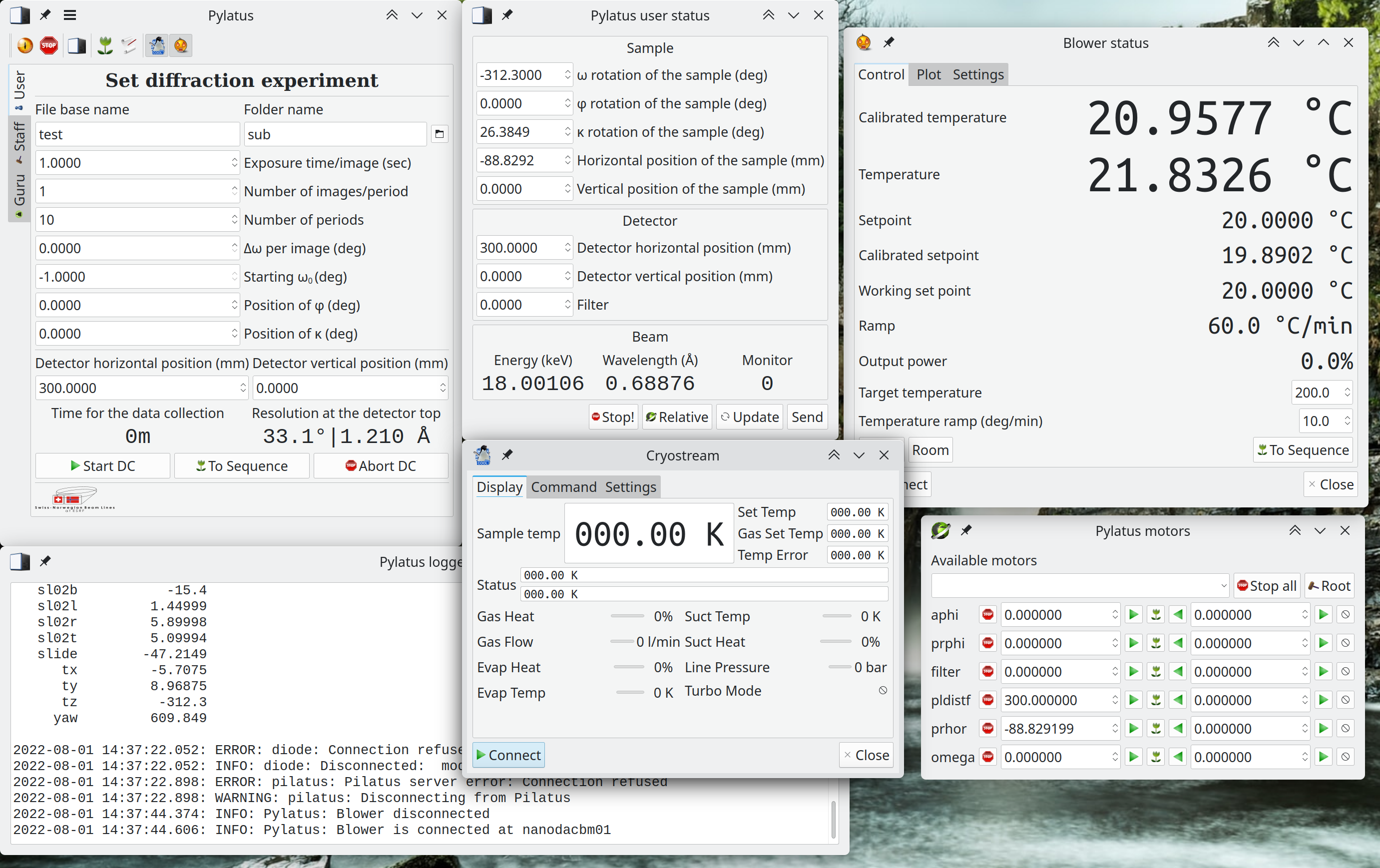
Pylatus beamlines
Currently, Pylatus is working at the following ESRF beamlines:
- BM01 --- Swiss-Norwegian CRG beamline (diffraction)
- BM31 --- Swiss-Norwegian CRG beamline (diffraction and EXAFS)
- ID28 --- Side station of the Inelastic Scattering beamline
- BM20 --- The Rossendorf CRG Beamline
Why Pylatus?
Pylatus is trying to combine a simple and easy-to-learn interface together with complexity required for modern synchrotron experiments. User can collect individual or multiple datasets for single crystals or powders using GUI but more advanced options for complicated data collection are also available (see scripting in Python).
Pylatus gives total control over beamline starting up from the optics (mirrors and monochromators) down to the sample environment. It includes a lot of batteries, such as:
- Kappa or chi goniometer for single crystal and powder experiments. Infinite spinner is also available.
- All motor views and controls, moving motors relatively and absolutely.
- Pseudo-motors (such as energy, wavelength or grouped motions).
- Continuous scans (1D or meshes) for all motors using as counters:
- any SPEC counters;
- Silicon diodes connected to nano-ammeters (currently, only Keithley 6485 is supported);
- Pilatus detector itself (summing up all pixels on the frame using AVX512, AVX or SSE4 intrinsics if available);
- Beamline auto-alignment (using routine scenarios, like scanning a motor and positioning it to the intensity maximum value);
- Automatic beamstop search on the measured frames (using AVX512, AVX or SSE4 intrinsics if available; calibration required).
- Automatic storing experimental values (temperature, pressure, voltage, etc.) into the headers of the output CBF files.
- Powerful scripting in Python for complex experimental scenarios and sequences.
- Native control of the sample environment devices, such as:
- Eurotherm nanodac and 2408 for high temperatures;
- Oxford Cryostream devices for low temperatures;
- Lakeshore 3xx for helium temperatures;
- ISEG high voltage;
- Keithley 2200 low voltage;
- Implementation of the ESRF data policy.
- Possibility to connect to SPEC sessions and run everything available from SPEC.
Pylatus source code
Pylatus is free and open source software (under GPLv3 license), the source code can be found in the Pylatus git repository.
Pylatus is mainly written in C++ but it has some modules written in pure C and Rust. Pylatus depends on Qt5 (min version 5.12). Pylatus contains additional daemons, such as:
- dcubroker - a daemon running on the Dectris DCU. It collects cbf files from the camserver and sends them to Pylatus (written in Rust).
- icepapd - a daemon running the ESRF ICEPAP crates (written in Rust).
Cite Pylatus
If you use Pylatus or its parts, please cite this paper: A new multipurpose diffractometer PILATUS@SNBL, J. Synchrotron Rad. (2016). 23, 825-829 .
Pylatus scripting API
Pylatus API Objects
Technical details
Pylatus is dynamically linked against a local version of Python, but only Python>=3.6 is supported.
If you do not want to use any Python scripting in Pylatus, it should be compiled with -DNO_PYTHON=1.
Thus, Python in Pylatus is the same interpreter which is installed in your system, and you can import any module which is installed for you system interpreter, i.e. this would work
import numpy as np
if you have numpy installed. Pylatus extends the system modules with a few objects specific for a typical diffraction experiment.
It is important to note that the Pylatus do not run the script immediately but creates a sequence of actions which can be inspected by user and run after.
diffractometer
The main object which allows to script diffraction scan. (Diffractometer script source code)
In Pylatus Script editor the diffractometer object is already imported into Python interpreter, but if you
have external .py files, then you can import it as:
from pylatus_diffractometer import PyDiffractometer
diffractometer = PyDiffractometer()
Macros example can be found in here: https://git.3lp.cx/dyadkin/bm01macros/src/branch/master/lab6.py
Instance methods:
- scan
- sleep
- abort
- setPhiScan
- setOmegaScan
- align
- setDet (BM31 only)
scan
diffractometer.scan(
*,
folder=None,
filename=None,
nperiods=None,
nframes=None,
exposure=None,
start=None,
step=None,
omega=None,
dOmega=None,
omegaEnd=None,
phi=None,
phiEnd=None,
kappa=None,
kappaEnd=None,
pldistf=None,
pldistd=None,
plrot=None,
plvert=None,
nop=False
)
Creates a data collection sequence action.
This method accepts only the keyword arguments. If an argument is not set, its value is taken from the main
Pylatus window, from the "Set diffraction experiment" widget (excluding nop argument).
The method parameters are the following:
folder: string which specifies the directory where the data will be stored;filename: string with file base name;exposure: exposure time in seconds, float number;nframes: number of frames, integer number;nperiods: number of periods, integer number;start: starting value for scanning motor (synonymous foromegafor an abstract beamline)step: step value for scan (synonymous fordOmegaandangleStepfor an abstract beamline)dOmega: angular step in degrees per image foromegaaxis, float number.omega: starting angle in degrees for omega axis when omega-scan is set or just omega position if phi-scan is set, float number;omegaEnd: end omega angle in degrees for psi scan, float number;phi: starting angle in degrees for phi axis when phi-scan is set or just phi positionphiEnd: end phi angle in degrees for psi scan, float number; if omega-scan is set, float number;kappa: kappa angle in degrees, float number;kappaEnd: end kappa angle in degrees for psi scan, float number;pldistd: position of the detector inside the frame in mm, float number;pldistf: position of the frame in mm, float number;plrot: detector angle in degrees, float number;plvert: vertical offset of the detector in mm, float number;nop: bool, stands for no periods: ifFalsethefilenameparameter will be extended with the period number, i.e. filename_0001p.cbf; ifTruejustfilenamewill be used, i.e. filename.cbf.
sleep
diffractometer.sleep(seconds)
Creates a sequence action to sleep: diffractometer does not do any action for a number of seconds.
abort
diffractometer.abort()
Creates a sequence action to abort the current diffraction data collection. Useful when data are collected periodically and user wants to stop it automatically when the temperature ramp is done.
setPhiScan
diffractometer.setPhiScan()
Creates a sequence action to set phi axis as the main scanning axis.
In this case, the angular step will be called dPhi, since 2021-04-01
or dOmega for older version.
Attention: if you switch omega and phi scans in the script, you HAVE TO specify
both omega= and phi= parameters in the diffractometer.scan method.
setPsiScan
diffractometer.setPsiScan()
Creates a sequence action to set psi axis as the main scanning axis.
setOmegaScan
diffractometer.setOmegaScan()
Creates a sequence action to set omega axis as the main scanning axis.
Attention: if you switch omega and phi scans in the script, you HAVE TO specify
both omega= and phi= parameters in the diffractometer.scan method.
align
diffractometer.align()
Create a sequence action for the diffractometer auto alignment procedure. All the values must be set in the Guru option Autoscan before using this method.
setDet (BM31 only)
diffractometer.setDetX(value)
diffractometer.setDetY(value)
diffractometer.setDetZ(value)
diffractometer.setDetR(value)
Sets GUI field for detector position in the Pylatus main window to the value.
Usage examples:
diffractometer.setOmegaScan()
diffractometer.scan(folder="omega_scan", phi=0, omega=-55, dOmega=1, nperiods=10)
diffractometer.setPhiScan()
diffractometer.scan(folder="phi_scan", phi=0, omega=-55, dPhi=1, nframes=5)
motor
motor("name")
The object which allows to script individual motor motions, where name is name of the motor.
(Motor script source code)
In Pylatus Script editor the motor object is already imported into Python interpreter, but if you
have external .py files, then you can import it as:
from auxygen_motors import PyMotors
motor = PyMotors()
Macros example https://git.3lp.cx/dyadkin/bm31macros/src/branch/master/mono.py
Instance methods:
move
motor("name").move(position, *, now=False)
creates a sequence action for the motor name to move it to the absolute position.
If now=True, then it does not create the sequence action but moves the motor immediately
(recommended only for very advanced users).
moveRelative
motor("name").moveRelative(position, *, now=False)
creates a sequence action for the motor name to move it to the relative position.
If now=True, then it does not create the sequence action but moves the motor immediately
(recommended only for very advanced users).
wait
motor.wait(*, now=False)
Creates a sequence action to wait until all the moving motors reach desirable positions.
If now=True blocks the script immediately and waits until all the motor finish
(recommended only for very advanced users).
It does not need a motor name.
position
motor('name').position() -> float
Returns current position of the motor name.
(recommended only for very advanced users)
isAt
motor('name').isAt(position) -> bool
Calculates whether the motor name is at the position taking into account the motor precision.
(recommended only for very advanced users)
Usage examples:
# move motors to the starting positions
motor('omega').move(0)
motor('phi').move(90)
motor('kappa').move(-55)
motor('gon1').move(-55)
# wait until all the motor reach starting positions
motor.wait()
# make a scan
diffractometer.scan()
cryostream
The object which operates the Cryostream gas blower. (Cryostream script source code)
In Pylatus Script editor the blower object is already imported into Python interpreter, but if you
have external .py files, then you can import it as:
from auxygen_cryostream import PyCryostream
cryostream = PyCryostream()
All commands have now keyword arguments, which runs immediately instead of creating a sequence action.
Default value now==False.
Instance methods:
cool
cryostream.cool(temperature, *, now=False)
Creates a sequence action to cool the cryostream to the temperature as quick as possible.
Temperature interval is from 80 K to 500 K (float).
ramp
cryostream.ramp(temperature, ramp_rate, *, now=False)
Creates a sequence action to ramp the cryostream to the temperature with the ramp rate of ramp_rate.
Temperature interval is from 80 K to 500 K (float), ramp rate is from 1 to 360 K/hour.
end
cryostream.end(ramp_rate, *, now=False)
Creates a sequence action to ramp the cryostream to the room temperature the ramp rate ramp_rate
and switches it off. Ramp rate is from 1 to 360 K/hour.
wait
cryostream.wait(*, now=False)
Creates a sequence action for cryostream to wait until it reaches the temperature specified by one of the previous commands.
setTurbo
cryostream.setTurbo(enable: bool, *, now=False)
Creates a sequence action for cryostream to enable (if enable == True) or disable (if enable == False)
the turbo mode.
temp
cryostream.temp()
Returns the current temperature.
blower
The object which operates the Eurotherm controlling a standard ESRF hot gas blower or the Hermann's heater. (Blower script source code)
In Pylatus Script editor the blower object is already imported into Python interpreter, but if you
have external .py files, then you can import it as:
from auxygen_blower import PyBlower
blower = PyBlower()
Instance methods:
ramp
blower.ramp(temperature, ramp_rate)
Creates a sequence action to ramp the blower to the temperature with the ramp rate of ramp_rate.
Temperature interval is from 20 C to 1000 C (float), ramp rate is from 1 to 1000 C/min.
wait
blower.wait(temperature=None)
Creates a sequence action to wait for the temperature specified in the blower.ramp.
If the optional parameter temperature (float) is not None, then blower waits until temperature is reached.
For some samples (often damaged by x-rays) it is useful not to measure all the time but at certain temperatures without stopping the ramp, i.e.
blower.ramp(1000, 10)
for t in range(100, 1001, 10):
blower.wait(t)
diffractometer.scan()
Then we would measure diffraction every 10 C with permanent ramp.
lakeshore
The object which operates the Lakeshore temperature controller. (Lakeshore script source code)
In Pylatus Script editor the lakeshore object is already imported into Python interpreter, but if you
have external .py files, then you can import it as:
from auxygen_lakeshore import PyLakeshore
lakeshore = PyLakeshore()
Instance methods:
setpoint
lakeshore.setpoint(output, target)
Creates a sequence action to set target temperature (in K) for the lakeshore at the output (usually 1 or 2).
ramp
lakeshore.ramp(output, state, value)
Creates a sequence action to set value temperature ramp (in K/min) for the lakeshore at the output
(usually 1 or 2) depending on the state value which can be False (switch off ramp) or True (switch on ramp).
range
lakeshore.range(output, range)
Creates a sequence action to set the heater range at the output.
range can be either one of the following values:
0- Off1- Low2- Medium3- High
heater
lakeshore.heater(output, value)
Creates a sequence action to set the heater to manual value at the output.
value can be in a range from 0 to 100 %.
pid
lakeshore.pid(output, p, i, d)
Creates a sequence action to set p, i, d coefficients at the output.
wait
lakeshore.wait()
Creates a sequence action for the Lakeshore to wait for the temperature set with lakeshore.setpoint.
iseg
The object which operates the ISEG high voltage device. (Iseg script source code)
In Pylatus Script editor the iseg object is already imported into Python interpreter, but if you
have external .py files, then you can import it as:
from auxygen_iseg import PyIseg
iseg = PyIseg()
Instance methods:
setVoltage
iseg.setVoltage(voltage, ramp_rate)
Creates a sequence action to set the voltage value with the ramp_rate.
Maximum voltage depends on the model (usually 2000 V).
Ramp rate is from 2 to 255 V/s.
ramp
iseg.ramp(voltage, ramp_rate)
Alias for iseg.setVoltage.
wait
iseg.wait()
Creates a sequence action to wait until the voltage is set by the previous command.
keithley
The object which operates the Keithley2200 voltage/current device. (Keithley script source code)
In Pylatus Script editor the keithley object is already imported into Python interpreter, but if you
have external .py files, then you can import it as:
from auxygen_keithley import PyKeithley
keithley = PyKeithley()
Instance methods:
setVoltage
keithley.setVoltage(voltage)
Creates a sequence action to set voltage value in V at the Keithley output.
setCurrent
keithley.setCurrent(current)
Creates a sequence action to set current value in A at the Keithley output.
stop
keithley.stop()
Creates a sequence action to switch off the Keithley output.
spec
The object which connects to a SPEC session and runs macros in there. (Spec script source code)
In Pylatus Script editor the spec object is already imported into Python interpreter, but if you
have external .py files, then you can import it as:
from auxygen_spec import PySpec
spec = PySpec()
Important: unlike other objects, spec does not create a sequence action but runs directly and immediately, i.e.
it changes the Pylatus script editor to a prompt, then the Python interpreter blocks until the spec command is
finished. If you need to run a spec macro as a sequence action, consider using to_sequence
decorator.
Macros example https://git.3lp.cx/dyadkin/bm31macros/src/branch/master/spec.py
Instance methods
__init__
You can create a spec connection as:
s = spec('host:session')
Since creation of the connection is expensive and time-consuming operation, Pylatus puts them in a cache for future reuse.
run
spec('bm31spec2:experiment').run('ct 1')
Runs a spec command immediately and blocks until it is finished. You can abort the command at any moment.
to_sequence
to_sequence is a decorator to put a block of Python code into a sequence action.
(to_sequence source code)
In Pylatus Script editor the to_sequence decorator is already imported into Python interpreter, but if you
have external .py files, then you can import it as:
from auxygen_seq import to_sequence
Usages example https://git.3lp.cx/dyadkin/bm01macros/src/branch/master/musst.py
Explanation
Imagine we have a function which connects to spec and runs a macro there. In our case, we have a spec
macro chan_on which turns on a channel chan on the MUSST card:
def musst_chan_on(chan):
spec('snbla2:rhmusst').run(f'chan_on {chan}')
If we call this function as is
musst_chan_on(8)
it will turn on TTL signal on our MUSST. But we may want to put our spec command into the sequence, then we could
write it like:
@to_sequence
def musst_chan_on(chan):
'''Musst turns channel 'chan' ON'''
spec('snbla2:rhmusst').run(f'chan_on {chan}')
After that, calling
musst_chan_on(8)
it will create a sequence action instead of running it immediately.
Format doc strings
You can use Qt format strings to add some info into sequence action:
@to_sequence
def musst_chan_on(chan, time):
'''Musst turns channel '%1' for %2 seconds'''
spec('snbla2:rhmusst').run(f'chan_on {chan}')
from time import sleep
sleep(time)
spec('snbla2:rhmusst').run(f'chan_off {chan}')
which will generate an action as following:
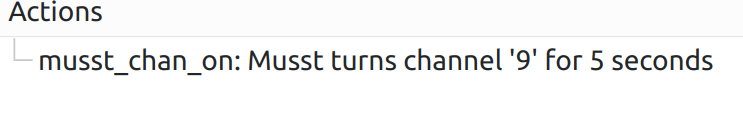
%1 will be substituted with the first argument value, %2 with the second one, etc., up to 9 arguments are allowed.
Limitations
Due to fragile substance of the decorator there are certain limitations on its usage:
- Only a top level function with only positional arguments can be decorated. Classes, methods, metaclasses, lambdas, closures or other smart thing will not work, i.e.
@to_sequence
class TooSmart: # wrong! classes cannot be decorated
pass
class EvenSmarter(TooSmart):
@to_sequence
def cool_method(self): # wrong! methods cannot be decorated
pass
@to_sequence
def silly_function():
'''I am a very silly function''' # ok!
@to_sequence
def silly_function(a=1): # wrong! keyword arguments are not allowed
'''I am another very silly function'''
@to_sequence
def smart_function():
'''I am a very smart function'''
@to_sequence
def silly_function(): # wrong! closures cannot be decorated
'''I am a very silly function'''
-
The decorated function can get any number of positional arguments (no keywords), but they must be either:
- strings,
- integer numbers,
- float numbers.
More complicated data structures (i.e. dicts, lists, other objects) will not work:
@to_sequence
def spec_hello(greet, num): # <--- ok!
'''Prints `greet` `num` times in spec'''
spec('bm31spec2:experiment').run(f'for (i=0; i<{num}; i++) print({greet});')
@to_sequence
def spec_run_all(list_of_commands): # <--- wrong! python list (or any Iterable) cannot be an argument
'''Runs list of commands in spec'''
for command in list_of_commands:
spec('bm31spec2:experiment').run(f'{command}')
- The decorated function must have a doc-string, which will be used as a part of the sequence action name.
@to_sequence
def silly_function():
'''I am a very silly function''' # ok!
@to_sequence
def dummy_function():
return None # wrong! there is no doc string
- The decorated function cannot capture anything from the outer scope, i.e. neither imports, nor variables, nor other functions or types:
from time import sleep
sleep_time = 5
@to_sequence
def spec_sleep1():
'''Block spec prompt for 5 seconds'''
sleep(sleep_time) # wrong! sleep and sleep_time cannot be captured from the outer scope
@to_sequence
def spec_sleep2(sleep_time):
'''Block spec prompt for `%1` seconds'''
from time import sleep
sleep(sleep_time) # ok, sleep is imported inside the function, sleep_time is an argument
If you find these limitations too strict, your patches eliminating them are always welcome.
log
The object which puts a message into Pylatus log window and, consequently, into Pylatus log file. (log script source code)
In Pylatus Script editor the log object is already imported into Python interpreter, but if you
have external .py files, then you can import it as:
from auxygen_log import PyLogger
log = PyLogger()
Important: unlike other objects, log does not create a sequence action but runs directly and immediately, i.e.
it changes the Pylatus script editor to a prompt, then the Python interpreter blocks until the spec command is
finished. If you need to run a log macro as a sequence action, consider using to_sequence
decorator.
Instance methods
info
log.info("Simple info message")
Prints an info message into Pylatus log window and, consequently, into Pylatus log file.
warn
log.warn("Warning message")
Prints a warning message into Pylatus log window and, consequently, into Pylatus log file.
error
log.error("Error message")
Prints an error message into Pylatus log window and, consequently, into Pylatus log file.
brainboxes
The object which operates the remote IO BrainBoxes devices. (BrainBoxes script source code)
In Pylatus Script editor the brainboxes object is already imported into Python interpreter, but if it is going
to be called from an external .py file, then the manual import is needed:
from auxygen_brainboxes import PyBrainBoxes
brainboxes = PyBrainBoxes()
Instance methods:
setInput
brainboxes.setInput(address, channel, value, *, timeout=0, now=False)
Creates a sequence action (if now==False) which sets the input channel of a BrainBoxes device with address
to value.
The value equals to 0 is interpreted as off, anything else is on.
If timeout is not 0, then the input will be set to the inverted value after timeout milliseconds.
If now==True, the sequence action is not created, but the command is run immediately (no sequence action mode).
Useful Pylatus script snippets
- Measure all LaB6 patterns at once
- Measurements with Cryostream
- Eurotherm based device
- Modulation Enhanced Diffraction
- Measure the full sphere with Arinax micro-kappa
- Fast change of the mono energy for BM01
Measure all LaB6 patterns at once
dists = [0, 100, 200, 300, 400, 500]
heights = [-80, 0]
for d in dists:
heights = heights[::-1]
for h in heights:
diffractometer.scan(filename=f'lab6_{d:03d}_{h:02d}',
pldistf=d, plvert=h, omega=0,
dOmega=360, nperiods=1,
exposure=60, nframes=1, nop=1)
Measurements with Cryostream
# from 300 K to 200 K with a step 10
# and from 200 K to 80 K with a step 1 K
temps = list(range(300, 200, -10)) + list(range(200, 79, -1))
for t in temps:
# ramp with rate 360 K/hour
cryostream.ramp(t, 360)
cryostream.wait()
# sometimes it's useful to wait for temperature stabilization
diffractometer.sleep(120)
# Folder pattern is like Sample1_100K
diffractometer.scan(folder=f'Sample1_{t:03d}K')
Eurotherm based device (gas heat blower or Hermann's heater)
# from 400.5 K to 500.5 K with a step 0.5
# It is not possible to use range with float,
# Thus we import arange from numpy
from numpy import arange
temps = arange(400.5, 501, 0.5)
for t in temps:
# ramp 10 C/min
blower.ramp(t, 10)
blower.wait()
# Directory pattern is like Sample1_400.5C
diffractometer.scan(folder=f'Sample1_{t:.2f}C')
Modulation Enhanced Diffraction
import numpy as np
amax = 250 # max temperature amplitude
amin = 200 # min temperature amplitude
n = 50 # number of points
filename = 'name'
func = np.cos # use cosine function
points = np.linspace(0, 2 * np.pi, n)
temps = (amax + amin) / 2 + func(points) * (amax - amin) / 2
for i, t in enumerate(temps):
cryostream.ramp(t, 360)
cryostream.wait()
# nop=True means the filename pattern will be without _0001p
# part, i.e. name_001.cbf instead of name_001_0001p.cbf
diffractometer.scan(filename=f'{filename}_{i:03d}', nop=True)
Measure the full sphere with Arinax micro-kappa
dir = 'test'
runs = {
'run0': {'akappa': 0, 'aphi': 0, 'prhor': -44.5813, 'samtx': 1.6822, 'samty': 0.8764},
'run1': {'akappa': 90, 'aphi': 0, 'prhor': -44.3563, 'samtx': 1.9445, 'samty': 1.3063},
'run2': {'akappa': 90, 'aphi': 90, 'prhor': -44.3064, 'samtx': 1.8516, 'samty': 0.7455},
'run3': {'akappa': 90, 'aphi': 180, 'prhor': -44.6463, 'samtx': 1.3820, 'samty': 0.8077},
}
for r in runs:
motors = runs[r]
for m in motors:
motor(m).move(motors[m])
motor('omega').move(0)
motor.wait()
diffractometer.scan(folder=f'{dir}/{r}', kappa=motors['akappa'], phi=motors['aphi'])
Fast change of the mono energy for BM01
Fast energy change
from bm01macros import *
move_mono(<value>)
Possible values:
- PHEHR: parallel high energy beam (20 keV) with harmonic rejection;
from bm01macros import *
move_mono(PHEHR)
- PHE: parallel high energy beam (20 keV) with possible harmonics;
from bm01macros import *
move_mono(PHE)
- FHEHR: focused high energy beam (17 keV) with harmonic rejection;
from bm01macros import *
move_mono(FHEHR)
- FHE: focused high energy beam (17 keV) with possible harmonics;
from bm01macros import *
move_mono(FHE)
- PLEHR: parallel low energy beam (12 keV) with harmonic rejection;
from bm01macros import *
move_mono(PLEHR)
- PLE: parallel low energy beam (12 keV) with possible harmonics;
from bm01macros import *
move_mono(PLE)
- FLEHR: focused low energy beam (12 keV) with harmonic rejection;
from bm01macros import *
move_mono(FLEHR)
- FLE: focused low energy beam (12 keV) with possible harmonics;
from bm01macros import *
move_mono(FLE)
SNBL Toolbox

SNBL Toolbox is a set of useful programs for Pilatus data. If you use this program please cite this paper: A new multipurpose diffractometer PILATUS@SNBL
SNBL Toolbox is open source software covered by the GPLv3 license.
Download
To run the SNBL Toolbox in Windows you may need to install the MS Visual C++ Redistributable for Visual Studio 2015-2022. Usually you would prefer the version with avx enabled.
The Linux version of the SNBL Toolbox is compiled in Ubuntu 20.04 LTS.
The macOS version of the SNBL Toolbox is compiled in Sequoia-15 producing fat binaries for both (x86_64 and arm64) architectures.
Source code
Source code is freely available at the git repository https://git.3lp.cx/dyadkin/snbltb
Changelog
-
2025.10.30 - v2.4.0:
- Fix file sorting issue with file names not following the snbl naming strategy
- Fiewer: fix an improper cake view if radial and azimuthal bins are different (works with bubble>=4.3)
- Fiewer: Better powder patterns for cakes (works with bubble>=4.3) #22
- HeadEx: fix crashes when it reads a directory with no files #20
- Switch to Qt6
- Fat binaries for macOS
-
2024.6.25 - v2.3.9:
- If period number has more than 4 digits, Convert, Sleuth, HeadEx and Fiewer correctly sort those files
-
2024.6.7 - v2.3.8:
- Crysis: if template par file is specified, Crysis takes the wavelength from the CBF files anyway #15.
- Crysis: set d2 par value using 2theta value from CBF.
- Fiewer: fix crashes when mouse is moved while playing multi-frame images #16.
- Fiewer: show the beam center for CBF.
- Sleuth: fix possible crashes if integrating limits cross the frame border.
-
2023.3.1 - v2.3.7:
- Crysis: fix regression when Crysis does not create all the needed files for Crysalis.
- Fiewer: now can open Crysalis IMG files.
- HeadEx: support for Voltage CBF header key.
-
2023.1.25 - v2.3.6:
- Converter: fix incorrect periodical binning when it is started not from the very first period #14.
- Fiewer: fix incorrect numbers in Bubble cakes in Windows a0a374e4e6.
-
2022.8.3 - v2.3.5:
- Fiewer correctly displays detector gaps for Dectris Eiger.
- Fiewer can open Eiger frames from ID11@ESRF.
- Fiewer: fixed an issue when the shown image is one more pixel stretched than it should be.
- Fiewer can sum up frames in multiframe files (usually, Eiger HDF5) and export them as one EDF.
- HeadEx can now extract the Pressure header key.
-
2022.4.20 - v2.3.4:
- Fix regression in Crysis (since v2.2.4): Crysis creates a not containing mask file for Pilatus detectors.
- Converter now uses AVX and AVX2 CPU instructions which may stop Toolbox working on old computers.
-
2022.1.26 - v2.3.3:
-
2022.1.7 - v2.3.2:
- Fix Fiewer crashes when a Bubble cake is not square #11.
- Fiewer can open pyFAI generated HDF5 files.
- HeadEx: extract header values from EDF files.
-
2021.11.26 - v2.3.1
- Converter: fix incorrect summing of angle increments for periodical binnings.
- Converter: fix a possible crash if user deletes files from the list for conversion.
-
2021.10.28 - v2.3.0
- Fiewer shows cakes produces by Bubble bubbles #7.
- Converter: improve the speed of file scanning when adding an enormous amount of files.
-
2021.6.29 - v2.2.6
- Fiewer watches for newly appearing files (even on Windows)
- Fiewer can open tiff files with int32 values (usually Pilatus tiff)
- HeadEx can extract calibrated blower temperature: use Blower_calibrated in the list
-
2021.1.27 - v2.2.5
- Fix regression in Converter: very slow file scanning
-
2020.12.16 - v2.2.4
- Crysis: fix multiruns with KuMa-like kappa goniometer
- Converter: CBF 'Count_cutoff' header key is summed up while binning
- Fiewer: fix remembering to many arrow key clicks while listing heavy frames
-
2020.10.8 - v2.2.3
- Converter: Gandolfi-like binning for multi-temperature single crystal scans
- Fiewer: display angular information for CBF frames
-
2020.9.1 - v2.2.2
- Converter: fix incorrect binning of CBF frames periodically on Windows and MacOS
-
2020.8.26 - v2.2.1
- Converter: fix incorrect binning of CBF frames
-
2020.5.17 - v2.2.0
- Fiewer: new frame formats:
- Agilent esperanto, raw frames and packed with AGI_BITFIELD;
- TIFF;
- Dectris Eiger HDF5;
- Fiewer: fix various memory leaks happened when playing frames back and forward
- Fiewer: smart file sorting
- Fiewer: new frame formats:
-
2020.5.17 - v2.1.0
-
A new tool: Fiewer - the frame viewer. Fiewer is supposed to be a fast tool to show images. It is able to jump between frame files or frames inside a multiframe file (using tool buttons or keyboard arrow keys) sorting files in lexical order. Fiewer support frame types such as:
- EDF: all primitive types, multiframed files. Packed (gzip, etc) EDF format is not supported;
- CBF from Dectris Pilatus;
- Bruker format;
- Mar (non-spiral);
- JungFrau: only if master header in HDF5 format exists;
- Fit2D masks;
-
Converter: fix naming scheme while binning periodical datasets
-
-
2020.2.21 - v2.0.10
- Converter: properly sum up the header keys for merged frames
-
2019.4.15 - v2.0.9 (a9c76dfce9c6ac07789341fef61d902b01aa8071)
- Converter: fix wrong MD5 sum in merged CBF files, then ALBULA can open CBF files after the Toolbox again
- Converter: set negative intensities to the requested value even for one converted frame
- Converter: check the flux value for zero, otherwise a CBF header could have a 'nan' value for flux
-
2019.4.8 - v2.0.8 (70c41a5636f91804c299395e658cc7c344ad299f)
- Fix wrong update message
- Minor internal fixes for performance
-
2019.1.21 - v2.0.6 (ea5930cd008a856a6bbbeaa9dfbd1a51bb6b4454)
- SNBL Toolbox is rewritten in C++ and changes version from 1 to 2
- Crysis accepts a par file as a template for all processed directories
- To start from scratch, Crysis can remove all old files processed by Crysalis before. Use attentively!
- Converter can normalize the final frame on the Flux counter or the median value
- Various small fixes
-
2018.2.27
- 3DV: fix Windows bugs
- HeadEx: fix progress bar
- HeadEx: fix not working stop button
-
2018.1.16 (7b5973dbceb9)
- HeadEx can return CBF time as Unix time (number of seconds since January 1, 1970)
- HeadEx reads cbf files using multiple threads
-
2018.1.15 (6219065f6a4b)
- Fix various crashes in HeadEx
- New look for HeadEx
-
2017.12.14 (c302bcd79ef6)
- Crysis and Converter do not crash if there is an OS error (read-only filesystem, for example)
- 3DV does not create layers with just zeros on Windows
- SNBL Toolbox checks the updates from https://soft.snbl.eu/
-
2017.8.23 (344b86e56f28)
- Comply with new cryio API.
- Since August 24, 2017 Pylatus changes the position of the beam center to comply not with Fit2D (as before) but with Albula and pyFAI. Thus, crysis can now correctly handle the old and the new data.
- crysis shows the whole progress now.
Old SNBL Toolbox versions
-
2018.2.27:
-
2018.1.16:
-
2018.1.15:
-
2017.12.14:
-
2017.8.23
Bubble
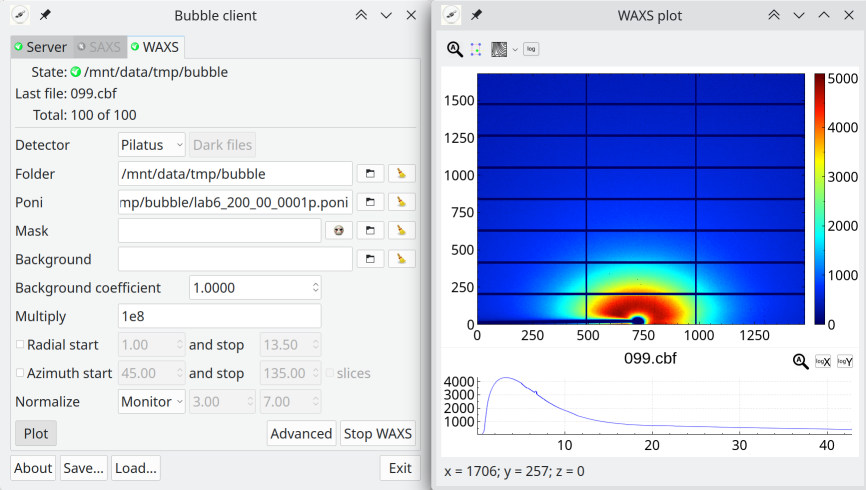
Bubble is a program for fast on-line powder integration for small and wide angle diffraction.
If you use this program please cite this paper: A new multipurpose diffractometer PILATUS@SNBL.
Bubble is open source software covered by the GPLv3 license.
Download
To run the Bubble client in Windows you may need to install the MS Visual C++ Redistributable for Visual Studio 2015-2022. Prefer AVX/SSE-enabled version.
The Linux version of bubble is compiled in Ubuntu 20.04 LTS.
The macOS version of bubble is compiled in Sequoia-15 producing fat binaries for both (x86_64 and arm64) architectures.
Source code
Source code is freely available at the git repositories:
- Full meta-package: https://git.3lp.cx/dyadkin/bubble.meta
- Client in C++: https://git.3lp.cx/dyadkin/bubblec
- Server in Rust: https://git.3lp.cx/dyadkin/bubbles
Changelog
-
2025.11.17 - v4.3.2:
- Fix issue when bubble mask is incorrectly saved on Windows #8.
-
2025.11.13 - v4.3.1:
- Fix issue when the detector correction window does not store attenuation and thickness parameters.
-
2025.10.30 - v4.3.0:
- Absolute path for integrated data: if the subdir parameter in the Server page is absolute (starts with / in unix or drive: in windows (c:, d:, etc.)), bubbles will store the data in there, recreating the needed directory structure #23
- bubbles may not receive notification from the OS if it is watching a network file system (mainly nfs), to fix this a --poll (-p) cli flag should be used #24
- Additional corrections for the effective sensor layer thickness and absorption #25
- Fix incorrect cake view in Fiewer #26, #29
- An option to switch off the tth integration when chi is requested #27
- Store chi and cake integrations in separate sub-directories #28
- Cosmic rays removal for integrated frames #30
- Option to inflate gaps for a number of pixels #32
- Fix incorrect negative division for flood field #30
- Remove Frelon/Pilatus detector distinction #31
- Fix averaging of hf5 files when all the data is written into the same dat file #35
-
2023.1.27 - v4.2.2:
- Lexicographical sorting of the azimuthal slices.
- bubbles: new CLI argument (--poll, -p) to poll watched directories periodically; it is useful for network shares (like NFS) when bubbles does get INotify events from kernel.
-
2022.1.7 - v4.2.0:
- Client does not crash anymore, if an empty mask is applied (bubblec #4).
- Client can disconnect from server from now (bubblec #5).
- Fix SNBL azimuthal range (bubbles #18).
- Auto mask Eiger gaps (bubbles #19).
- Average is now possible for multiframed data (i.e. Eiger) (bubbles #20). Averaging cannot go through frame border.
- If user stops integration, averaging also stops (bubbles #21).
-
2021.10.27 - v4.1.0:
- Nice cake view (can be seen via SNBL Toolbox Fiewer) (bubbles #7).
- Bubble should now guess the pixel size from a detector name from new PONI format.
-
2021.7.23 - v4.0.2:
- Fix server crash when a user specifies radial range above the bin limit (bubbles #15).
- Fix for server which could not open certain unlucky EDF files (bubbles #16).
-
2021.7.16 - v4.0.1:
- Fix memory leaks in server (bubbles #13).
- Add FReLoN detector in the PONI parser.
-
2021.5.27 - v4.0.0:
- Dectris Eiger HDF5 with bitshuffle filter support (cryiorust #3).
- Cakes are now saved in EDF to avoid rounding problems with CBF format. Use SNBL Toolbox::Fiewer to look at them (bubble.meta #1).
- Dark background and sample files do not swap anymore (bubblec #3).
- SAXS and WAXS calibration factors are not swapped anymore (bubblec #2).
- The plot is now properly updated with the latest integrated file (bubblec #1).
- Averaging is not so greedy for memory anymore (bubbles #10).
- Previously, switching normalization from Monitor to None did not have any effect, it is fixed now (bubbles #8).
- On linux bubbles may create files available only to its user (i.e. 0600), it is fixed now (bubbles #2).
- Fix for not reading symlinks in bubbles (bubbles #1).
- Bubbles is using Actix framework now (bubbles #5).
- Fix for overflows in some Mar files.
- Add support for tif files with 32 and 64 signed integers (mainly tested with files produced by Dectris Pilatus).
-
2020.5.17 - v3.1.0:
- Add the 'None' beamline: the images are shown and integrated 'as is' without applying any transformations. This beamline type has to be chosen in case if the data are not measured neither on SNBL-BM01@ESRF nor Dubble@ESRF.
- When both radial and azimuthal ranges were applied, the final integration pattern could have been calculated incorrectly. Fixed.
- Fix crashes of the client on linux.
- Save mask ROI separately in the client.
- Save mask in Fit2D format. Then it can be reused in Fit2D or pyFAI. The beamline should be set to
Nonethen. - When an integrated 2D pattern (or a mask image) is zoomed enough then the pixel intensity is shown for all the visible pixels.
-
2020.2.7 - v3.0.0:
- The server is rewritten in the Rust programming language. The integration speed increased up to 20% and the memory consumption decreased about two times.
- On-line integration is now available on the all three main platforms, monitor and photo counters and transmission are correctly taken into account.
- Integrator can now average files by a number given by user. The averaging happens per directory. Contradicting issues between the on-line integration and the averaging are quite expected and cannot be avoided.
- In the client it is again possible to set logarithmic scale for the axes in 2D plots.
- The server can run as a normal UNIX daemon (forking, logging, etc).
- Minor usability fixes in the client.
- Qt updated to v5.14.1
-
2019.7.30 - v2.0.0:
- The client is rewritten in C++.
- New widgets for 1D and 2D plots. To rescale the values for 2D plots, please, use the mouse wheel and dragging on the colorbar. The right button allows to change the color scheme.
- The mask is now plotted using polygon lines (like in Fit2D), double click finishes the mask shape. The mask can be dragged or removed (via the context menu).
- The issue, when a few overlapping polygons produce gaps in the final mask, has been resolved.
- Issues with discontinuity for azimuthal integration has been solved for angles 0, 90, 180, 270. The discontinuity for angle 360 still exists.
- The client does not crash anymore when the number of bins set to 1.
- The button with the server settings has been removed. Partially, those settings migrated to the advanced options, separately for SAXS and WAXS.
-
2018.11.28
- Bugfix: fix multicolumn files.
-
2018.11.8.1
- Support for new poni files from pyFAI-0.16.
- Bugfix: saving mask with 'less' or 'more' values does not crash the client after reopen.
- Bugfix: when WAXS is not normalized, the background was not properly subtracted.
-
2018.7.9
- Hopefully, the final fix of crashes in Windows.
- New mask: the saved mask file contains everything inside: both an image and ROIs which can be edited after load.
-
2018.6.21
- Fix various Windows related bugs.
-
2018.6.8
- Fix various client crashes.
-
2018.6.7
- Fixing various bugs in the Windows version;
- Fixing bugs with spline correction;
- Correct calculation of the error bars for patterns normalized by the background;
- bubbleg can be run with option
-resize N, where is a number to bin images byNxNpixels; it decreases the image size which are to be transferred over the network. By default,N=1in Linux andN=4in Windows and macOS.
-
2018.3.7
- Fixing issues with ellipsoidal mask in the client;
- Bruker images;
- Distortion correction for Frelon.
-
2018.2.22
-
New integration server written in Go, thus, the features are not yet supported:
- Spline files for Frelon-like cameras;
- Brueker images;
If you need these features, please use an older version. Everything else should work as expected.
-
Bubble client shows the image after integration, which means, if radial or azimuthal limits are applied, then they will also be seen in the 2D plot;
-
Now it is possible to set both numbers of radial and azimuthal bins in the Options page, thus, bubble can save radially integrated patterns as a function of 2θ (Q), azimuthally integrated patterns as a function of χ and 2D-Cake images in CBF format;
-
Recursive integration: the bubble server integrates in the parent directory and in all the internal directories;
-
There is no need anymore to do start/stop, if the directory is the same: bubble not just waits for new files, but it also checks if the older files have changed and integrates them accordingly.
-
Mar Image Plate image support;
-
-
2017.10.22 (513b8fb114c6)
- Bruker image support.
-
2017.10.21 (7c24e8753ce8)
- Updated UI.
- Fine integration tuning on the server tab:
- Number of bins;
- Polarization correction;
- Solid angle correction;
- Double precision.
- Client checks updates.
- Minor bug fixes.
-
2017.8.24 (70ff9f8e8d97)
- If normalization is done not by monitor, then the Bubble does read the Flux value from the CBF header.
- The options 'Speed' and 'Super speed' are applied on-line: there is no need to restart the integration process.
- Minor bug fixes.
Previous versions of Bubble
-
2020.5.17 - v3.1.0:
-
2020.2.6 - v3.0.0:
-
2019.7.30 - v2.0.0:
-
2018.11.28:
-
2018.11.8.1:
-
2018.7.9:
-
2018.6.21:
-
2018.6.8:
-
2018.6.7
-
2018.3.7
-
2018.2.22
-
2017.10.23 (513b8fb114c6)
-
2017.10.21 (7c24e8753ce8)
-
2017.8.24 (70ff9f8e8d97)
Medved
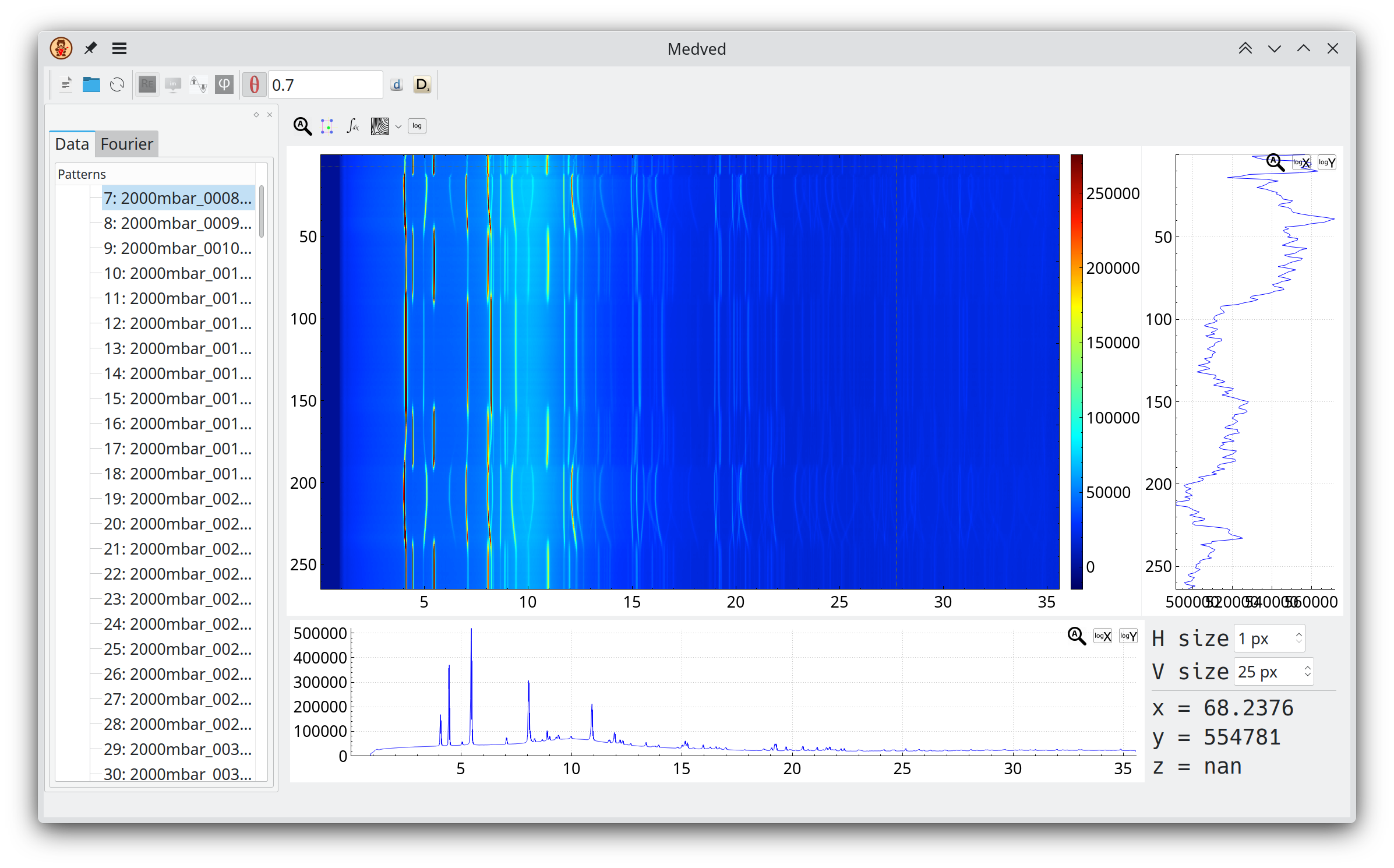
Medved (Modulation-Enhanced Diffraction Viewer and EDitor) is software that allows a user to load a number of powder patterns (standard two- or three-column ASCII data files) or hkl files (in SHELX format), inspect them separately or together, calculate Fourier transforms (using fast Fourier transform algorithms) and inspect separate harmonics as well as their real or imaginary parts.
If you use this program please cite this paper: Frequency analysis for modulation-enhanced powder diffraction.
Medved is open source software covered by the GPLv3 license.
Download
To run Medved in Windows you may need to install the MS Visual C++ Redistributable for Visual Studio 2015-2022.
The Linux version of Medved is compiled in Ubuntu 20.04 LTS.
The macOS version of Medved is compiled in Sequoia-15 producing fat binaries for both (x86_64 and arm64) architectures.
Source code
Medved source code is freely available at the git repository:
Changelog
-
2025.10.30 - v2.4.0:
- Fix file sorting issue with file names not following snbl naming strategy
- Switch to Qt6
- Fat binaries for macOS
-
2024.6.25 - v2.3.1:
- Correct sorting if number of periods in a filename contains more than four digits
-
2023.3.1 - v2.3.0:
- Save Fourier data as text files.
-
2021.9.17 - v2.2.0:
- The crosshair size can be adjusted to cover the actually integrated area (View->Adjust crosshair size).
- Export vertical cut and 2d images.
- Fit vertical cut with pseudo-Voigt function (Process->Fit...).
- Fourier is calculated only on-demand (i.e. when the Fourier tab is switched).
-
2020.5.17 - v2.1.0:
- Medved opens patterns in parallel.
- If the directory with data contained non pattern files, they might have appeared in the data dock. Fixed.
- When the image is zoomed enough, Medved shows the pixel value for the all visible pixels.
-
2020.1.14 - v2.0.0:
- Major version change to 2
- The core is rewritten in C++, the performance is significantly boosted
- Reload button
- Auto-reload option (only in Linux and macOS), Medved automatically loads newly created patterns
- Changeable size of the cuts
-
2017.11.20.1 (d5fbc19b6f0e)
- New UI
- Fix update checking
- Fix for the crash when a new dataset is loaded
-
2017.9.12.1 (401e64c7806c)
- Minor bug fixes
-
2017.9.12 (88aaf6d3b2e4)
- MEDVED checks updates at https://soft.snbl.eu
- New UI based on standard Qt widgets
- Fourier calculations are done n times faster now, where n is a number of loaded files
- Fixing the About window: new website; the links are clickable now
- Fourier view correctly shows plot ticks when switches something->phase->something
- Refactoring of the code
- Minor bug fixes
-
2016.12.2 (a807bc7b7cc8)
- Porting to Qt5
- Small improvements
Previous versions
-
2017.11.20.1
-
2017.9.12.1
-
2017.9.12
-
2016.12.2
Chirok
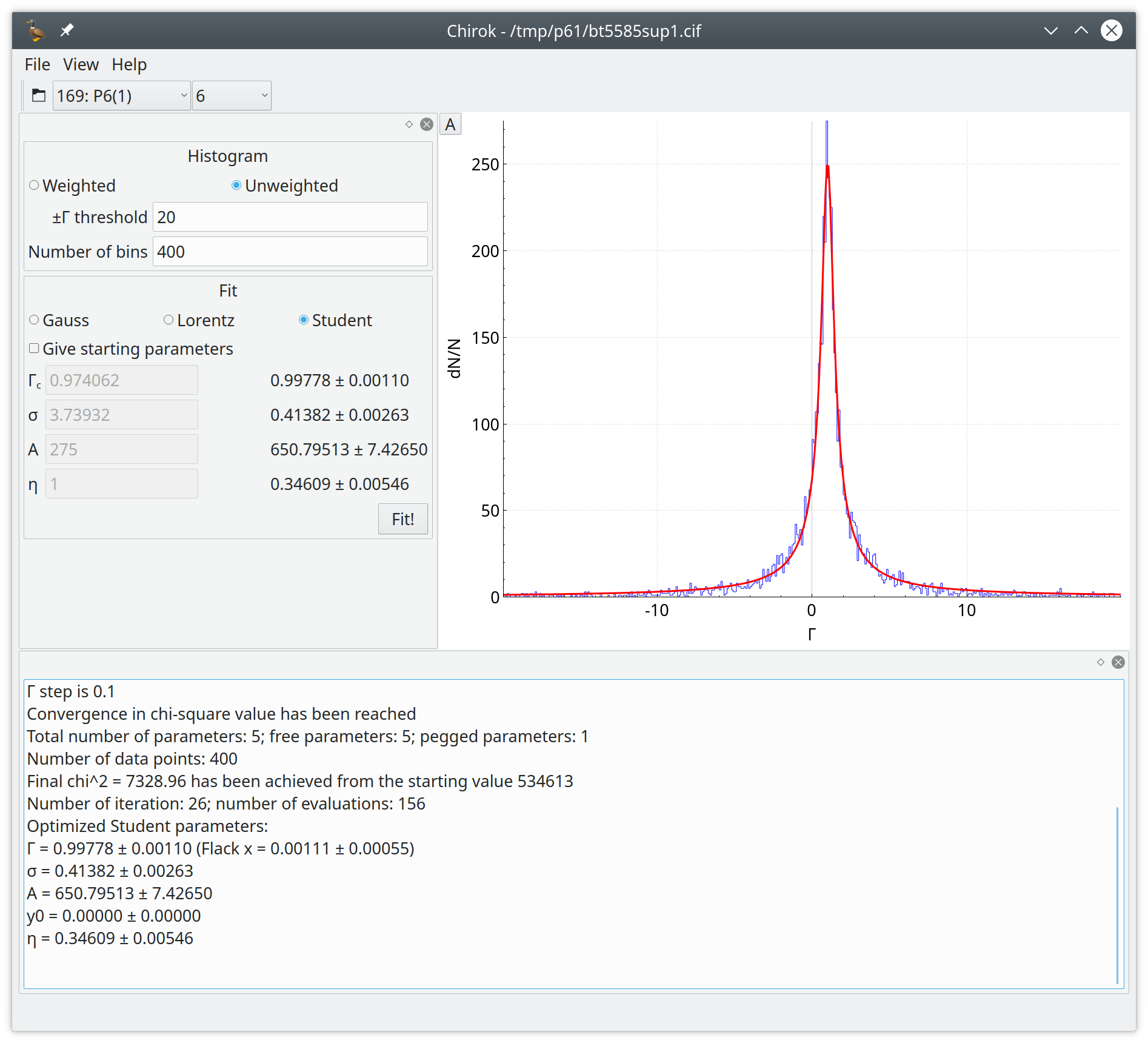
Chirok is a program for statistical analysis of the ratio of calculated and measured Parsons quotients.
Chirok is open source software covered by the GPLv3 license.
If you use this program, please cite the following paper: Chirok: a post-refinement tool to analyse absolute structure.
Download
To run Chirok in Windows you may need to install the MS Visual C++ Redistributable for Visual Studio 2015-2019.
The Linux version of Chirok is compiled in Ubuntu 16.04, thus it should work in any rather new distributions.
Source code
Source code is freely available at the git repository: https://git.3lp.cx/dyadkin/chirok
Changelog
-
2020.7.28 - v1.1.2:
- Add paper into the about windows.
-
2020.5.20 - v1.1.1:
- Minor fixes.
-
2020.2.28 - v1.1.0:
- Chirok predicts the Gamma threshold and the number of bins using the Random Forest algorithm.
- Remove funny characters from the log output on Windows.
- Fix minor issues with zooming and autoscaling.
-
2019.7.24 - v1.0.0:
- First release.
-
2018.12.13 - v0.1.0:
- Initial alpha release.
Previous versions
GIuSAXS: GISAXS and GIWAXS data viewer
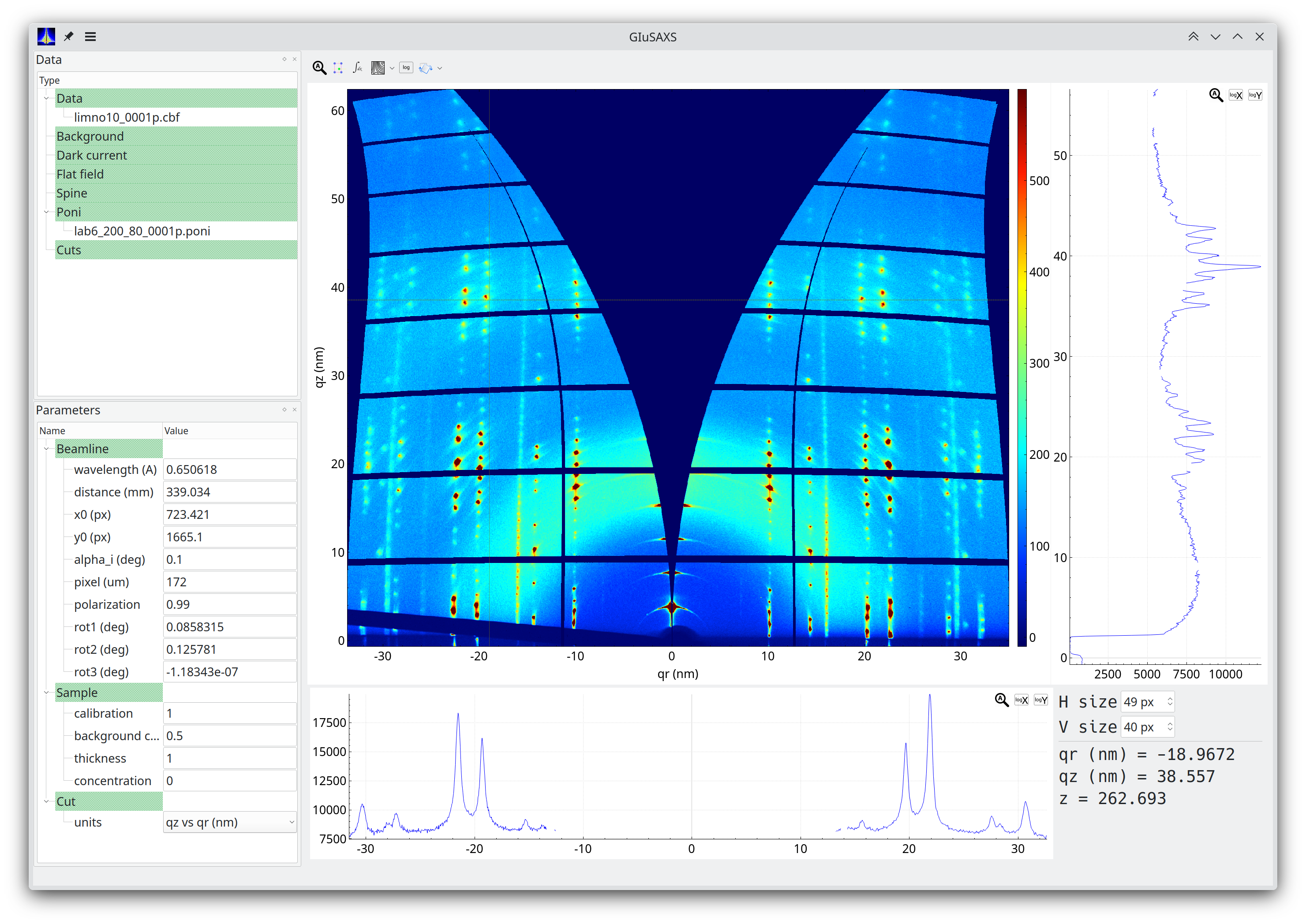
GIuSAXS is a tool to inspect GISAXS and GIWAXS data. The program is made in collaboration between:
- Giuseppe Portale, Zernike Institute for advanced Materials at the University of Groningen;
- Daniel Hermida Merino, Dubble@ESRF;
- Eduardo Solano, ALBA;
- Vadim Diadkin, SNBL@ESRF.
GIuSAXS is open source software covered by the GPLv3 license. GIuSAXS is funded by Dubble@ESRF.
Download
To run GIuSAXS in Windows you may need to install the MS Visual C++ Redistributable for Visual Studio 2015-2019.
The Linux version of the GIuSAXS is compiled in Ubuntu 16.04 LTS, thus it should work in any rather new distributions.
Source code:
Source code is freely available at the git repository: https://git.3lp.cx/dyadkin/giusaxs.
Changelog
-
2023.1.31 - v0.2.1:
- Export processed data as EDF.
-
2022.1.7 - v0.2.0:
-
2021.11.12 - v0.1.0:
- Reading a PONI file for geometry calculations.
- Ewald sphere corrections for qz vs qr.
- Sequential calculation of the cuts and storing them in ASCII files.
- Calculation of transmission.
-
2021.3.10 - v0.0.1
- Initial release of the version, rewritten in C++.
-
2017.10.23 (24a6613848e0)
- Support for Bruker images.
-
2017.10.22 (4793fe9afb8b)
- View menu to switch back the docks.
- Minor fixes.
-
2017.4.29 (dc1272d9cf36)
- Initial release.
Previous versions
-
2017.10.23 (24a6613848e0)
-
2017.10.22 (4793fe9afb8b)
-
2017.4.29 (dc1272d9cf36)
seq_shel: Sequential run of Crysalis and shelx
seq_shel allows you to run Crysalis and shelx sequentially for multiple single crystal datasets.
Better description can be found in the ECM Poster.
seq_shel is open source software covered by the GPLv3 license.
Download
Source code
Source code is freely available at the git repository:
Previous versions
SRO calculator
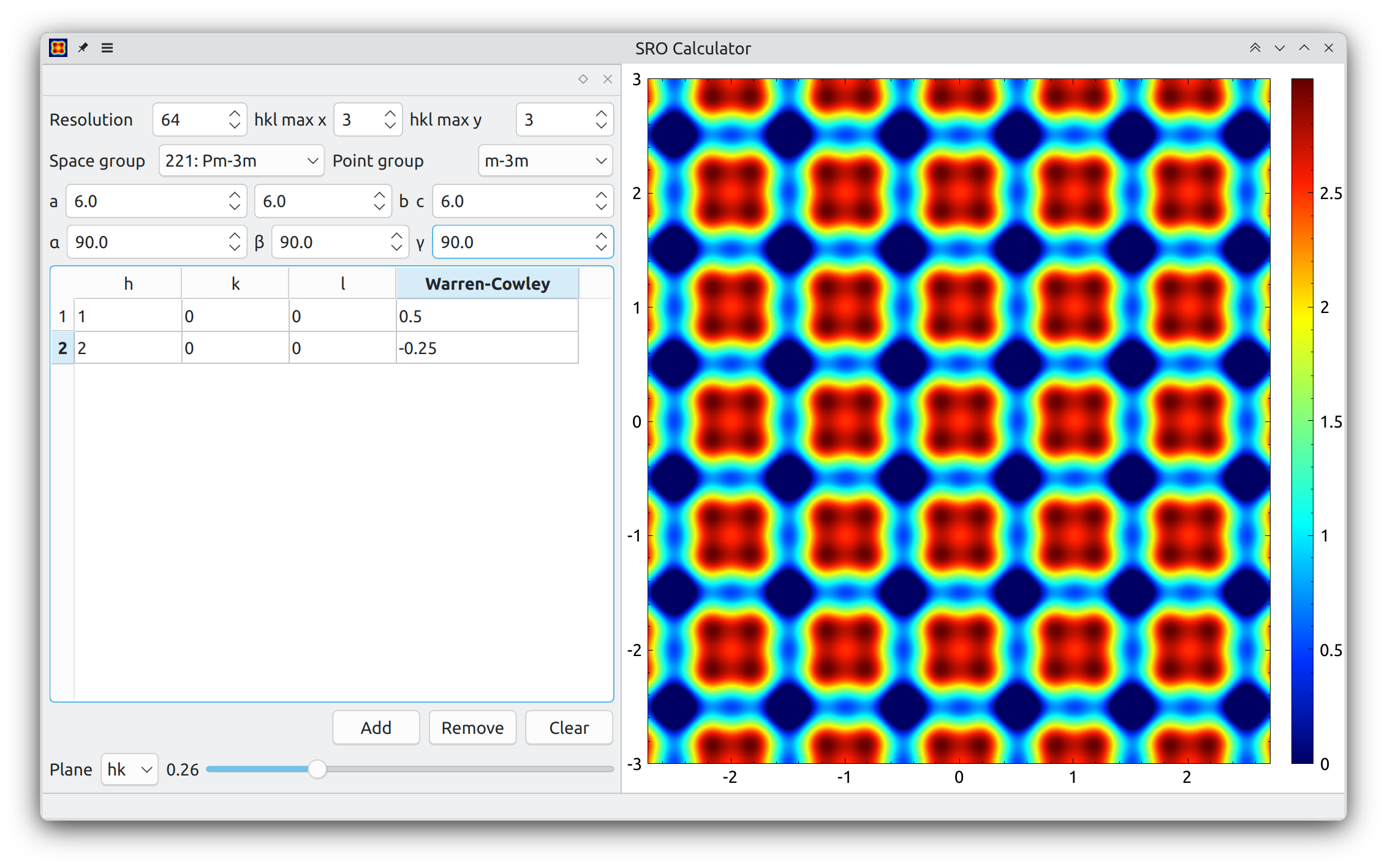
Short range correlations calculator.
SROc is open source software covered by the GPLv3 license.
Download
On Windows to run the SNBL Toolbox you may need to install the MS Visual C++ Redistributable for Visual Studio 2015-2022.
The Linux version of the SROc is compiled in Ubuntu 20.04 LTS.
The MacOS build contains executables for the both x86_64 and arm64 architectures.
Source code
Source code is freely available at the git repository https://git.3lp.cx/dyadkin/sroc
Changelog
-
2025.9.23 - v1.1.0:
- Change hkl to uvw
- Add about widget
-
2025.9.22 - v1.0.0:
- Initial release.
